 ERIK BENSON & BJORN HOGBERG. ATTRIBUTION 4.0 INTERNATIONAL (CC BY 4.0)
ERIK BENSON & BJORN HOGBERG. ATTRIBUTION 4.0 INTERNATIONAL (CC BY 4.0)
DNA—the biological information-storage unit and the mechanism by which traits are passed on from generation to generation—is more than just an essential molecule of life. In the chemical sense, the nucleic acid has properties that make it useful for nonbiological applications. Researchers are now using DNA to store massive amounts of data, for example, including books and images, a Shakespearean sonnet, and even a computer operating system, with data encoded in the molecule’s nucleotide sequences. At an even more fundamental level, DNA is a critical building block of nanoscale shapes and structures. Researchers have created myriad nanoscale objects and devices using the nucleic acid, with applications in biosensing, drug delivery, biomolecular analysis, and molecular computation, to name but a few. DNA provides a highly specific route to building nanostructures. While the field is still addressing how to scale...
DNA as bricks and mortar
 DNA AS BRICKS AND MORTAR: The straightforward and consistent pairing of DNA’s nucleotide bases make the molecule a reliable building material. Depending on the sequence, DNA strands can crossover to adjacent helices, creating a branch point.
DNA AS BRICKS AND MORTAR: The straightforward and consistent pairing of DNA’s nucleotide bases make the molecule a reliable building material. Depending on the sequence, DNA strands can crossover to adjacent helices, creating a branch point.
See full infographic: WEB | PDF© ISTOCK.COM/SHUOSHUThe four canonical nucleobases—adenine, guanine, thymine, and cytosine—are inherently programmable, as adenine always pairs with thymine, and guanine with cytosine. Just as these bases encode the biological instructions for building and maintaining an organism, so too do they form the basic code of designing shapes using DNA. Two strands that have complementary sequences of these nucleotides can bind to each other to form a double helix structure with a diameter of about 2 nanometers, and a single turn of DNA (a helical pitch) measuring about 3.4–3.6 nm. This helix is quite stiff within lengths of 15 helical pitches, or about 50 nm (its “persistence length”), allowing DNA to be used as a rigid construction material.
While entire chromosomes are composed of tightly coiled DNA, the double helix itself is inherently linear, extending in only one direction. For it to be useful for construction in two or three dimensions, branched DNA junctions are created by the reciprocal exchange of strands. This phenomenon occurs in nature—for example, during formation of the Holliday junction, an intermediate in genetic recombination. Synthetic DNA sequences can be designed to pair in certain ways, resulting in branched junctions with helices that extend in more than one direction. DNA can also hold other bits of DNA together: sticky ends, or short, single-stranded overhangs at the tips of a helical nucleic acid, can be designed to bind to one another by proper sequence complementarity. Such sticky ends act as structural glue to bind DNA motifs or complexes, resulting in hierarchical self-assembly of these macromolecules into larger arrays and larger complex architectures. (See illustration below.)
Crystallographer Ned Seeman, now at New York University, first proposed the idea of using DNA to build molecular scaffolds in the early 1980s.1 Seeman, then at the State University of New York at Albany, grew frustrated trying to crystallize molecules the traditional way, by exploring a range of experimental conditions by trial and error. He hypothesized that branched DNA could be used to build a framework to solve this crystallization problem by holding the molecule of interest in a defined spatial position, effectively crystallizing it. In 1983, he designed DNA sequences that formed stable, immobile, branched junctions containing four arms, which would lay the foundation for designing higher-order DNA structures and arrays, including a designed three-dimensional DNA crystal.2
See “3-D Seer”
The study of DNA nanotechnology has expanded greatly in the past 35 years. Building upon the original four-arm DNA junction, Seeman’s group upped the number of arms possible in a single branched junction to 12. To achieve more robust assembly, Seeman’s group also designed a DNA motif called the double crossover, which contains two helical domains connected by two strand crossovers, and used these “tiles” to assemble the first two-dimensional crystalline lattice made out of DNA.3 Another example is the triple crossover motif, which contains three adjacent helices, each joined to its neighboring domain twice, which has also been used to assemble two-dimensional arrays. Such periodic arrays became a useful framework for hosting other molecules, as Seeman had originally intended, as well as in molecular computation and to study distance-dependent interactions between proteins.
Researchers have created myriad nanoscale objects and devices, with applications in biosensing, drug delivery, biomolecular analysis, and molecular computation, to name but a few.
Researchers have also used DNA to create nanoscale objects. In 1991, for example, Seeman’s group produced the first three-dimensional object made from DNA: a cube with double-helical edges. In 1994, the team made a truncated DNA octahedron with 14 faces. More recently, Chengde Mao’s group at Purdue University designed a three-point-star DNA motif and used it to assemble tetrahedra, dodecahedra, and buckyballs.4 Mao’s team also designed a five-point-star motif to create an icosahedron.
In all of these cases, researchers must create precise stoichiometric mixtures of purified DNA strands. The structures form as the mixtures are cooled, triggering the strands to hybridize to complementary regions of other strands—a process known as hierarchical self-assembly. This strategy is used to create a variety of nanoscale structures, with applications in sensing biological molecules, bioimaging, and drug delivery.
More recently, researchers have devised other techniques for building DNA-based architectures. Peng Yin’s group at Harvard University’s Wyss Institute developed alternate strategies to create large DNA nanostructures. In one approach, the researchers designed single-stranded DNA tiles that can attach to one another, with a full set of tiles forming a rectangular “molecular canvas.” They rendered desired two-dimensional shapes by selecting a subset of these single-stranded tiles, each of which acted as a pixel on the canvas. The investigators subsequently expanded on this strategy to create three-dimensional shapes and arrays using DNA bricks. But today, a majority of DNA nanostructures are based on yet another approach, known as DNA origami.
Welcome to the fold
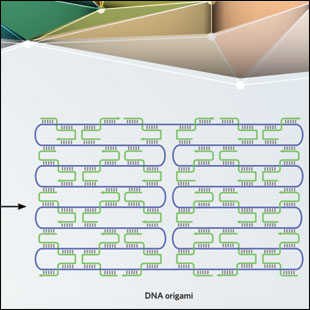 DNA ORIGAMI: By folding a long, single-stranded DNA scaffold using short, single-stranded “staples,” researchers can build any shape they like.
DNA ORIGAMI: By folding a long, single-stranded DNA scaffold using short, single-stranded “staples,” researchers can build any shape they like.
See full infographic: WEB | PDFARUN RICHARD CHANDRASEKARAN BUNNIES: ERIK BENSON & BJÖRN HÖGBERG. ATTRIBUTION 4.0 INTERNATIONAL (CC BY 4.0)In 2006, a smiley face made out of DNA appeared on the cover of Nature. In a study published in that issue, Paul Rothemund of Caltech described the process, dubbed DNA origami, that he used to create the smiley face, which measured approximately 100 nm in diameter and was imaged using an atomic force microscope (AFM). He began with a long piece of single-stranded DNA, called the scaffold strand, from the viral genome of the bacteriophage M13 (~7 kilobases). Then, with the addition of hundreds of short complementary oligonucleotides called staple strands, the scaffold was folded into the desired shape.5 As with hierarchical self-assembly, the structures were made using annealing procedures that involved heating and cooling the DNA mixture to facilitate binding of the short staple strands to different regions of the scaffold.
This strategy is largely dependent on the sequence and length of the single-stranded scaffold, thus limiting the size of the origami structure created. However, the ease of preparation and the flexibility of designing a variety of two- and three-dimensional structures has made DNA origami a go-to technique in the field. In addition, this method does not require purified DNA strands or exact stoichiometric mixtures of component strands.
Over the past decade, researchers have optimized the technique for higher yields, greater design complexity, and easier purification of desired structures, introducing isothermal annealing procedures, in which the temperature stays constant, for construction (instead of the traditional heating and cooling down of oligonucleotide mixtures). William Shih’s group at the Wyss Institute extended the concept of DNA origami to create the first three-dimensional objects from pleated sheets or bundles of DNA helices. The architectures made using origami are much larger than the DNA objects assembled from DNA tiles. Hao Yan’s group at Arizona State University produced DNA objects with curved surfaces such as hollow spheres and flasks, demonstrating the versatility of designing complex structures using DNA. Another notable example is the DNA origami box, developed by the groups of Kurt Gothelf and Jørgen Kjems of the Centre for DNA Nanotechnology at Aarhus University, Denmark. This box has a lid that could be unlocked by external DNA strands.
In 2015, Yan’s group constructed complex wireframe architectures using DNA origami, creating shapes and contours similar to a freehand drawing. In contrast to conventional DNA origami structures that contain tightly packed helices, Yan’s design allowed the creation of porous structures with a precision that is not attainable using traditional techniques, such as lithography. Such structures are potentially useful for the construction of delivery vehicles and fuel cells. They also used this strategy to create finite quasi-crystalline patterns, which remains a challenging task using other fabrication methods. That same year, Björn Högberg’s group at the Karolinska Institute in Sweden reported an automated procedure to create similarly complex shapes using origami. They wrote a computer algorithm that can provide the folding pathway for the scaffold strand to create any desired shape in a mesh-like pattern. (See illustration above.)
See “More Stable DNA Origami”
The design of such complex structures using DNA origami is supported by computer programs such as cadnano and CanDo, which can help users create any pattern and analyze resulting shapes. Most DNA origami construction is based on the M13 genome as the scaffold strand. Researchers have also used other single-stranded scaffolds, ranging in length from just 700 bases to some 50,000 bases, to create origami structures from tens to hundreds of nanometers in size. Recently, Seeman’s group designed a cross-shaped origami tile and created the first two-dimensional origami crystalline arrays, extending DNA origami’s dimensions into the micrometer scale.6 Single-stranded M13 DNA is now commercially available from companies such as New England Biolabs. Moreover, companies such as Tilibit now provide customers with a few options for scaffold strand lengths and also supply ready-made mixtures for prefabricated DNA origami designs.
Putting DNA nanostructures to work
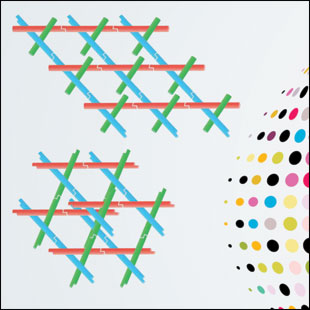 MULTIDIMENSIONAL DNA ARRAYS: Researchers can design small DNA motifs that self-assemble into lattices.
MULTIDIMENSIONAL DNA ARRAYS: Researchers can design small DNA motifs that self-assemble into lattices.
See full infographic: WEB | PDFARUN RICHARD CHANDRASEKARAN; © ISTOCK.COM/SHUOSHUSeeman’s original idea for using DNA as a building material was to spatially position “guest” molecules for the crystallization of hard-to-crystallize biomolecules. Over the years, researchers designed and assembled numerous DNA motifs into one- and two-dimensional arrays, and these have been used to host guests such as proteins, nanoparticles, antibodies, and quantum dots. But Seeman’s vision of using a DNA scaffold as a framework for macromolecular crystallization wasn’t fully realized until almost 30 years later, in 2009.
Seeman’s group used a DNA motif that spanned three dimensions, called a tensegrity triangle. The motif had threefold symmetry with three double-helical edges designed in an over-and-under fashion and six sticky ends, giving the structure the ability to connect to other tensegrity triangles. The assembly continues in an infinite arrangement, thus forming a crystal.7 (See illustration at right.) The cavities in this crystal provide space to accommodate guest molecules, thereby acting as a framework to obtain crystal structures. Seeman and his colleagues further showed that they could design such a DNA framework with bigger cavities, thus allowing researchers to encapsulate molecules of various sizes. In addition, the design could be asymmetric, allowing each edge of the DNA motif to have sequences for unique guests.
The fact that DNA structural scaffolds allow for the spatial positioning of proteins, specifically enzymes, allows researchers to use them for analyzing specific biomolecular interactions. For example, DNA motifs have been used to study the activities of a three-enzyme pathway—malate dehydrogenase/oxaloacetate decarboxylase/lactate dehydrogenase—with variable spatial distances and geometric arrangements. DNA scaffolds also allow researchers to increase the efficiency of enzyme cascades and to create synthetic enzyme pathways.
Beyond providing architectures that allow critical spatial positioning, DNA nanostructures can also be designed to respond to chemical cues, such as changes in temperature, pH, and ionic conditions. One of the most notable examples of such DNA nanodevices is a pH sensor called the I-switch, developed in 2009 by the University of Chicago’s Yamuna Krishnan, then at the National Centre for Biological Sciences in India.8 This device was based on the i-motif, a four-stranded structure containing stretches of cytosine repeat sequences that, under acidic conditions, pair with protonated cytosine (C+) instead of guanine. This pairing resulted in a conformational change from an open linear structure under physiological conditions (pH 7.3) to a closed triangular structure when pH dropped. In the switch, Krishnan and her colleagues included two fluorescent tags that only emitted light when brought into close proximity with each other by the acid-triggered shape change. The researchers used the I-switch to map spatial and temporal pH changes associated with endosome maturation in Drosophila blood cells and in C. elegans, demonstrating that DNA nanomachines could be used to measure real-time pH changes in living systems. Recently, the researchers developed another DNA nanodevice, called Clensor, to measure the activity and location of subcellular chloride channels and transporters in Drosophila blood cells.9
In addition to environmental stimuli, DNA constructs can be tailored to respond to more-specific signals, such as external DNA strands that bind specifically to a region of the nanostructure and induce conformational changes. Such biosensors are used for detection of nucleic acid or protein biomarkers and might one day aid in disease detection and treatment. One example is a DNA origami platform, developed by Seeman, that can detect single nucleotide polymorphisms. Researchers designed surface probes to bind to a target oligonucleotide, and if there is a mismatch between the probe and the target, the origami displays the alphabet letter—C, G, T or A—corresponding to the mismatched nucleotide when imaged using an AFM.10 DNA nanostructures tagged with aptamers—oligonucleotide sequences that bind to specific proteins, peptides, carbohydrates, and small molecules—can detect disease-related biomarkers and even cancer cells.
See “Antibody Alternatives”
DNA-based machines have also been developed to perform specific tasks. In 2009, Seeman’s group created a nanoscale assembly line: a DNA walker was able to pick up gold nanoparticle cargos from three different stations on a DNA origami platform. The assembly line can be designed so that the walker can pick up any of eight possible combinations of cargoes from the three stations. In 2014, another group of researchers created a different type of walker that was capable of transporting nanoparticle cargos along carbon nanotubes. And last year, Hendrik Dietz’s group at Technische Universität München in Germany used DNA origami components to create a rotary motor.11 Such innovations can be used to better understand the functioning of cellular machinery, such as the ATP synthase motor, as well as to create nanoscale devices that could potentially perform therapeutic functions in vivo.
See “DNA Machines Inch Forward”
Drug delivery and bioimaging
One of DNA’s main attractions is its biocompatibility. DNA has been used to build nanocages to house therapeutic cargoes, for example, and nanoscale robots functionalized with specific biomarkers can direct these structures to specific locations in the body, where they release their cargo upon binding to specific molecules. These constructs can be used to encapsulate fluorescent dyes, antitumor drugs, and peptide- and nucleic acid–based therapeutics such as siRNA and CpG sequences.
One of DNA’s main attractions is its biocompatibility.
One such example is a hexagonal barrel-shape DNA origami that encapsulated antibody fragments in its cargo binding sites.12 The two halves of this clamshell-like structure were connected at the rear by scaffold strand hinges. The front of the barrel contained a DNA aptamer “lock” that can be opened in the presence of the correct aptamer antigen (the “key”), thus providing a mechanism to release the encapsulated cargo upon reaching its target. One potential cargo is the well-known cancer therapeutic agent doxorubicin. Doxorubicin-loaded origami structures have been shown to effectively fight cancer both in human cell cultures and in mouse models. DNA origami triangular structures containing gold nanorods have also been designed to reach cancer cells in mouse models. Once at the target location, a near infrared (NIR) laser heats up the gold nanorods, thus killing the tumor cells—a process called photothermal ablation.
DNA nanostructures have also been useful for bioimaging purposes. Krishnan’s group, for example, has assembled a DNA icosahedron from five-arm junctions that can host cargoes within its cavity. The researchers encapsulated a fluorescent biopolymer within the icosahedron and used the complex to track cellular uptake pathways in Drosophila blood cells as well as in the whole organism C. elegans.13 The group used a similar strategy, but with quantum dots, to follow cellular uptake in HeLa cells. And last year, Chunhai Fan’s group at the Shanghai Institute of Applied Physics in China used tetrahedral DNA nanostructures with attached signaling peptides to track entry into cell nuclei.14 Such deliberate control over the entry and fate of DNA nanostructures in cells can spur the creation of more-efficient drug delivery pathways. While there are no DNA nanostructure–based drug delivery carriers yet in clinical trials, research towards this goal is being pursued.
DNA nanotechnology has grown out of its infancy and into adulthood. With the endless chemical strategies available to functionalize DNA strands, DNA nanostructures can host a variety of molecules and position them in predefined spaces to allow proximity-directed chemical reactions. As researchers continue to develop new techniques for building structures out of DNA, which can now be synthesized for as low as $0.001 per base pair, they will no doubt discover even more applications, in fields ranging from biology to medicine to biophysics.
Arun Richard Chandrasekaran is a senior scientist at Confer Health, Inc., which develops clinical-grade diagnostics for home use.
References
- N.C. Seeman, “Nucleic acid junctions and lattices,” J Theor Biol, 99:237-47, 1982.
- N.R. Kallenbach et al., “An immobile nucleic acid junction constructed from oligonucleotides,” Nature, 305:829-31, 1983.
- E. Winfree et al., “Design and self-assembly of two-dimensional DNA crystals,” Nature, 394:539-44, 1998.
- Y. He et al., “Hierarchical self-assembly of DNA into symmetric supramolecular polyhedra,” Nature, 452:198-201, 2008.
- P.W.K. Rothemund, “Folding DNA to create nanoscale shapes and patterns,” Nature, 440:297-302, 2006.
- W. Liu et al., “Crystalline two-dimensional DNA-origami arrays,” Angew Chem Int Ed, 50:264-67, 2011.
- J. Zheng et al., “From molecular to macroscopic via the rational design of a self-assembled 3D DNA crystal,” Nature, 461:74-77, 2009.
- S. Modi et al., “A DNA nanomachine that maps spatial and temporal pH changes inside living cells,” Nat Nanotech, 4:325-30, 2009.
- S. Saha et al., “A pH-independent DNA nanodevice for quantifying chloride transport in organelles of living cells,” Nat Nanotech, 10:645-51, 2015.
- H.K.K. Subramanian et al., “The label-free unambiguous detection and symbolic display of single nucleotide polymorphisms on DNA origami,” Nano Lett, 11:910-13, 2011.
- P. Ketterer et al., “Nanoscale rotary apparatus formed from tight-fitting 3D DNA components,” Sci Adv, 2:e1501209, 2016.
- S.M. Douglas et al., “A logic-gated nanorobot for targeted transport of molecular payloads,” Science, 335:831-34, 2012.
- D. Bhatia et al., “A synthetic icosahedral DNA-based host-cargo complex for functional in vivo imaging,” Nat Commun, 2:339, 2011.
- L. Liang et al., “Single-particle tracking and modulation of cell entry pathways of a tetrahedral DNA nanostructure in live cells,” Angew Chem Int Ed, 53:7745-50, 2014.
Interested in reading more?




